Microscopy data playbook
A suitable and flexible data management strategy is essential for effective and trustworthy science.
Our goal for data is to maximize access, understanding, analysis speed, and provenance while reducing barriers, unnecessary storage bloat, and cost.
Data perspectives
We think about data using three different perspectives:
- Level
- Origin
- Flow
Each perspective requires us to think through different considerations for storage, access, and provenance management. Managing microscopy data is related to other data types, with some nuance. For more details, see our previous article on data sharing practices for many different biological data types (including microscopy images)(Wilson et al. 2021).
1. Level
The data level indicates the stage and amount of bioinformatics processing applied. For example, the lowest data level, or “raw” data, are the images acquired by the microscope. (Technically, the biological substrate is the “rawest” data, but we consider the digitization of biological data to be the lowest level). Following the raw form, scientists apply various bioinformatics processing steps to generate various forms of intermediate data (see Figure 1).
With microscopy data, there are many different kinds of intermediate data; each typically of different sizes and thus have different storage and sharing requirements. Each intermediate data type has different requirements for storage and sharing.
2. Origin
Where data come from also requires unique management policies. Data can originate from within (either the lab or collaborators (both academic and industry)) or externally (data already in the public domain).
It is important to consider access requirements and restrictions, particularly when using collaborator data. For example, it is never ok to share identifiable patient data. When analyzing private data, we apply the same standards as public data, as it is helpful to remember that most data will eventually be in the public domain.
3. Flow
Besides the most raw form, data are dynamic and pluripotent; always awaiting new and improved processing capabilities. To determine short, mid, and long term storage solutions, we need to understand how each specific data level was processed at the specific moment in time (data provenance), and how each data level will ultimately be used.
We also need capabilities to quickly reprocess these data with new approaches. Consider each data processing step as a new research project, waiting for improvement.
Flow also refers to users and data demand. We need to consider data analysis activity at each particular moment. For example, if the data are actively being worked on, multiple people should have immediate access. We need to align data access demand with storage solutions and computability.
Microscopy storage solutions
We consider three categories of potential storage solutions for microscopy-associated data:
- Local storage
- Internal hard drive
- External hard drive
- Cloud storage
- Image Data Resource (IDR)
- Amazon/GC/Azure
- Figshare/Figshare+
- Zenodo
- Github/Github LFS
- DVC
- Local HPC
- One Drive/Dropbox/Google drive
- No storage
- Immediate deletion
Each storage solution has trade-offs in terms of longevity, access, usage speed, version control, size restrictions, and cost (Table 1).
| Solution | Longevity | Version control | Access | Usage speed | Size limits | Cost |
|---|---|---|---|---|---|---|
| Internal hard drive | Intermediate | No | Private | Instant | <= 18TB (Total) | ~$15 per TB one time cost |
| External hard drive | High | No | Private | Download | <= 18TB (Total) | ~$15 per TB one time cost |
| IDR | High | Yes | Public | Download | >= 2TB (Per dataset) | Free |
| AWS/GC/Azure | Low | Yes | Public/Private | Instant | >= 2TB (Per dataset) | $0.02 - $0.04 per GB / Month ($40 to $80 per month per 2TB dataset) |
| Figshare | High | Yes | Public | Download | 20GB (Total) | Free (Details) |
| Figshare+ | High | Yes | Public | Download | 250GB > x > 5TB (Per dataset) | $745 > x > $11,860 one time cost (Details) |
| Zenodo | High | Yes | Public | Download | >= 50GB (Per dataset) | Free (Details) |
| Github | High | Yes | Public/Private | Instant | >= 100MB (Per file) (Details) | Free |
| Github LFS | Intermediate | Yes | Public/Private | Instant | >= 2GB (up to 5GB for paid plans) | 50GB data pack for $5 per month (Details) |
| DVC | High | Yes | Public/Private | Download | None | Cost of linked service (AWS/Azure/GC) |
| One drive | Low | Yes | Public/Private | Instant | >= 5TB (Total) | Free to AMC |
| Dropbox | Low | Yes | Public/Private | Instant | Unlimited (Total) | $24 per user / month (Details) |
| Google drive | Low | Yes | Public/Private | Instant | >= 5TB (Total) | $25 per month (5 users)(Details) |
| Local cluster | Intermediate | No | Private | Instant | ||
| Immediate deletion | None | None | None | None | None | None |
Table 1: Tradeoffs and considerations for data storage solutions. Cost subject to change over time.
Microscopy data levels
From the raw microscopy image to intermediate data types including single cell and bulk embeddings, each data level has unique data storage and sharing considerations. We present a typical storage lifespan according to different data levels in Figure 1.
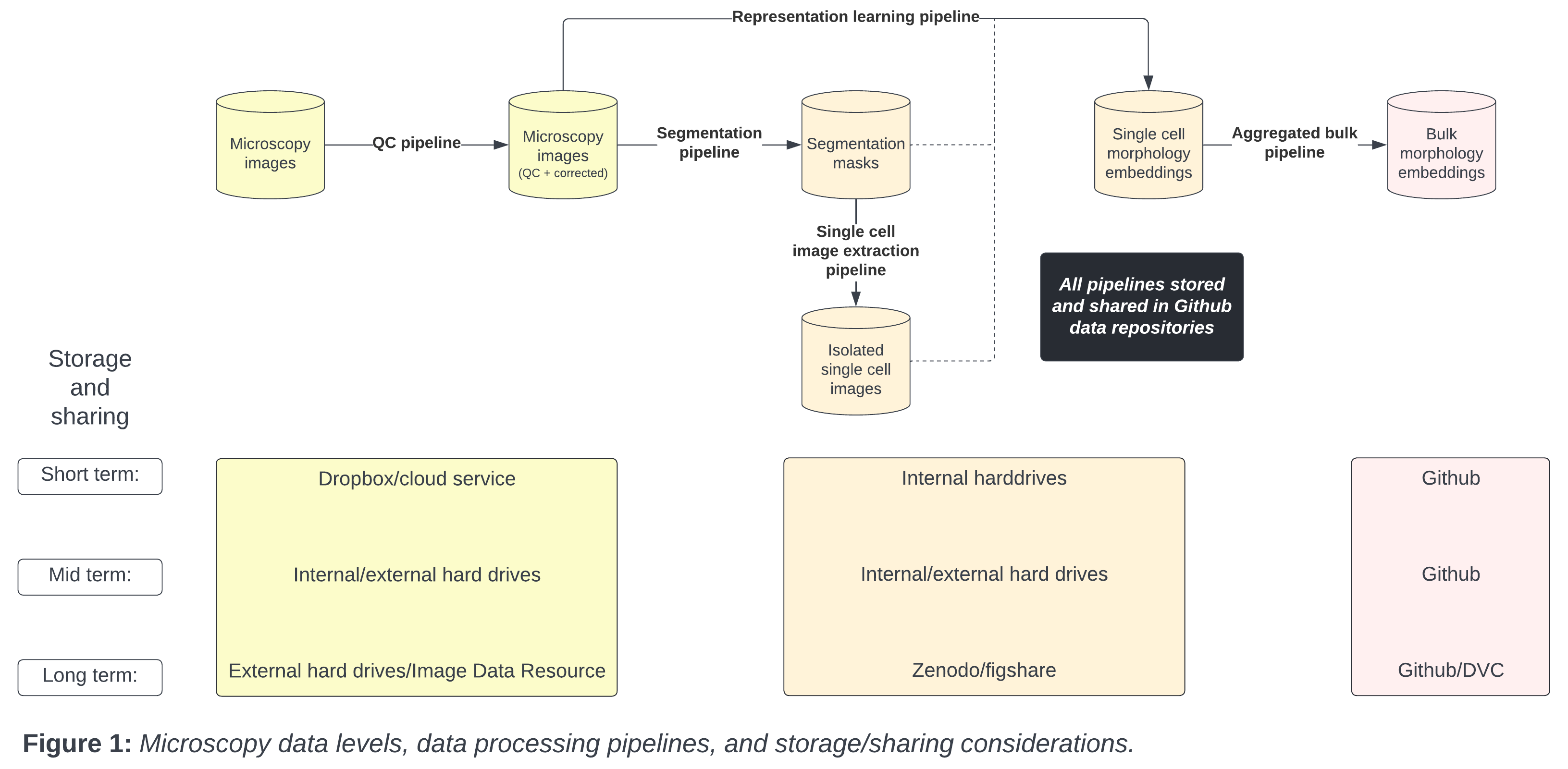
Metadata
Metadata for microscopy experiments have been discussed extensively, and are exceptionally important for data reproducibility and re-use.
For example, an entire Nature methods collection was recently devoted to microscopy metadata.
Most image-related metadata are stored alongside each image in .tiff formats, and many publicly available resources contain detailed instructions on how to access metadata.
This metadata must persist through the different data levels, and most often the metadata are small enough to store easily on github and local machines.
Previous post
Day 0 in the Way Lab
Next post
Illumination Correction Made Easier
